BioImaging and Optics Platform
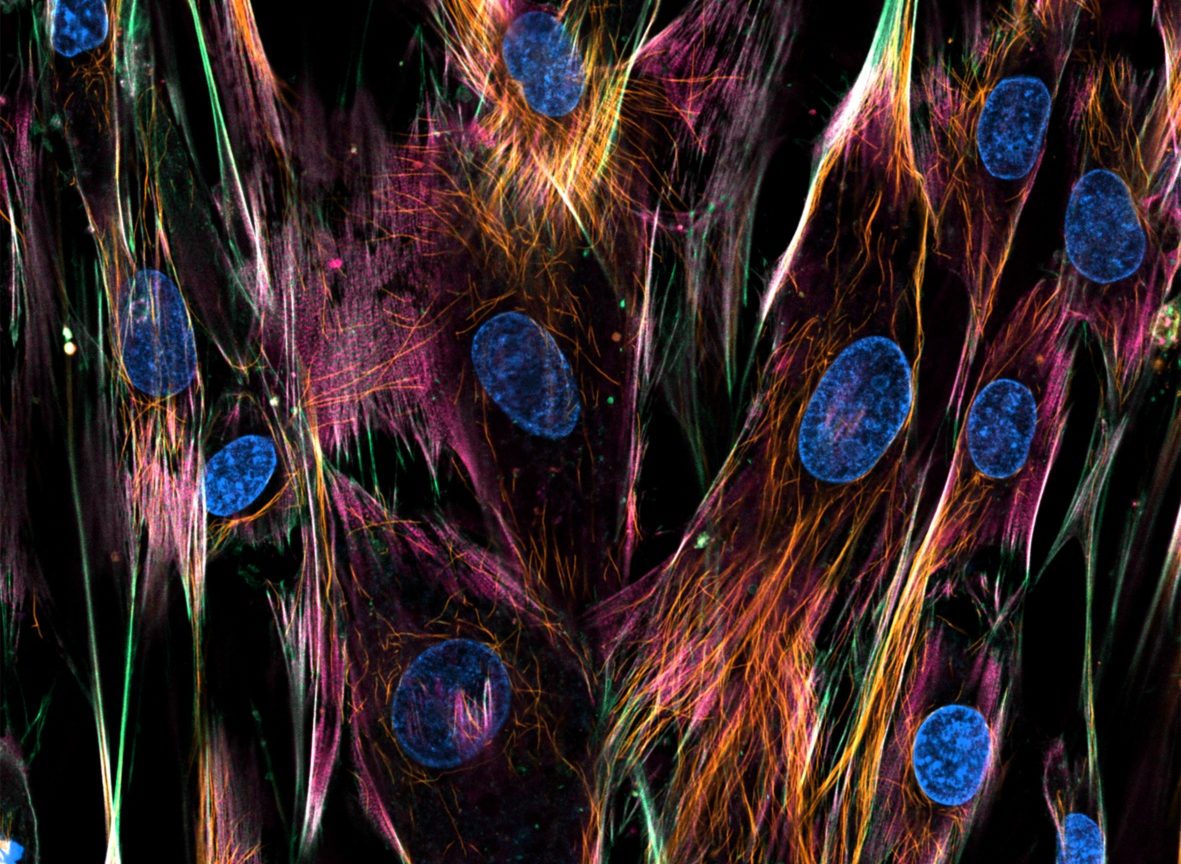
Fibroblasst with Nuclei, Microtubules, Actin and Mitochondria stainings, acquired on an LSM710 confocal microscope.
Our goal is to provide instruments and more importantly expertise to solve challenging (biological) questions with modern light-microscopy. Therefore the BIOP seeks to create a link between life sciences and engineering to investigate jointly these fundamental fields.
Upcoming Events
Highlights
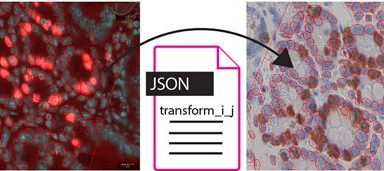
Whole Slide Registration Workflow Published
Our very own Nicolas has recently published a new article in Frontiers entitled: “An Open-Source Whole Slide Image Registration Workflow at Cellular Precision Using Fiji, QuPath and Elastix”
DEVILS is published in F1000R
Our Software Tool Article “DEVILS: a tool for the visualization of large datasets with a high dynamic range” is available on F1000R

ZIDAS 2023 is being co-organized by the BIOP
Switerland’s Image and Data Analysis School (ZIDAS) 2023 will be hosted by EPFL this summer!
Expertise

Microscopy
Widefield, Confocal, Spinning Disk, Lightsheet, Superresolution, Lifetime Imaging, Microdissection
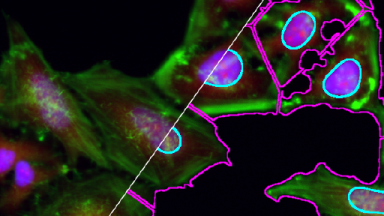
Image Processing
Workstations, Deconvolution, Deep Learning, Custom Workflows

Microscope Checkup Service
We are thrilled to introduce a new service aimed at ensuring the optimal performance of your light microscopes. To support your research and teaching efforts, we are pleased to announce the launch of our Microscope Checkup Service.